New method for analysing DNA
A new technique enables scientists to study genetic differences in cancer cells from the same tumour.
Analysing DNA has become a lot easier, as researchers can now analyse much larger pieces of genetic material than previously possible.
Gene sequencing normally involves decoding of the genetic code in separated DNA pieces of around 300 base pairs. The DNA pieces are then assembled into one big jigsaw puzzle.
But now researchers at the Technical University of Denmark (DTU) and Oxford University in England have developed a new technique that makes it possible to map the genetic patterns in DNA pieces of up to two million base pairs.
The new technique is even capable of extracting information from a single cell and then comparing its code with its neighbour cell. This could be interesting e.g. in cancer research.
”Cancer cells often experience a reorganisation of the genome, so that some DNA pieces are not located where they should be. There are also differences in the genetic structure of the individual cancer cells, depending on whether they are located on the inside or on the outside of the tumour,” says one of the researchers in the study, Jonas Nyvold Pedersen, of DTU Nanotech.
”With our new technique we can locate the misplaced DNA sequences and at the same time see the genetic differences in each individual cell. This could give us a deeper insight into the genetics of cancer.”
Fluorescent DNA is the key
One of the cornerstones of the new technique is a special DNA dye staining method, which had previously been developed at DTU.
During the dyeing process, the researchers use a fluorescent substance that binds between the individual bases in the DNA’s base pair. This lights up the entire DNA.
They then heat up the DNA to 76 degrees Celsius, at which point the bond between the adenine and the thymine bases (see Factbox) melts. The bonds between the guanine and the cytosine bases, however, stay intact as they have a higher melting point.
When the bond between adenine and thymine melts, the dye seeps out of the bond.
“When we cool down the DNA again, only the bonds between guanine and cytosine light up. We can then photograph the DNA and get a light intensity profile that gives us a rough indication of the composition of the base pairs,” says Pedersen.
A new chip was required
However, in order to decode the genetic information in the DNA, the researchers had to come up with another new technique.
The problem with DNA is that it behaves like a cluttered fishing line, which makes it impossible for the researchers to make heads and tails of it from only a picture.
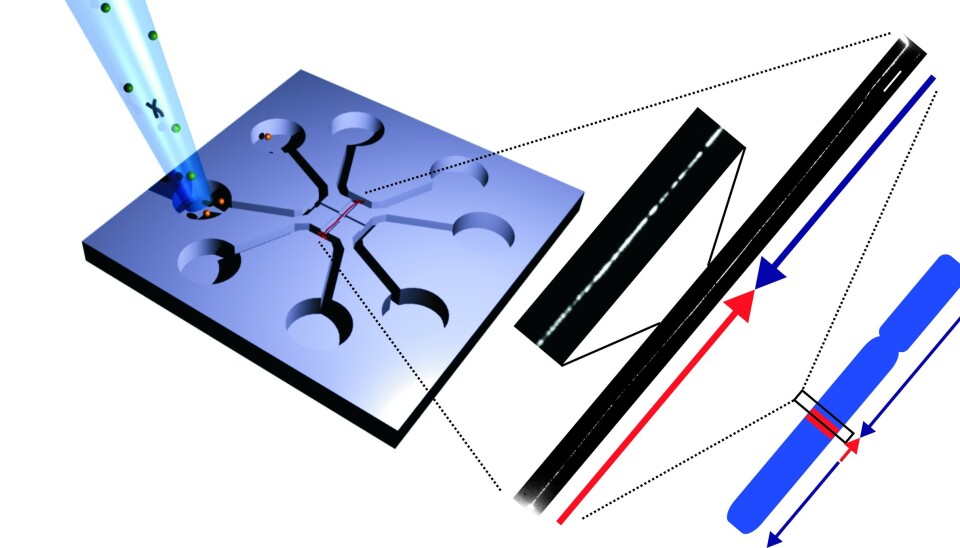
To solve this problem, Associate Professor Rodolphe Marie, also of DTU Nanotech, developed a ‘chip’ that could stretch the DNA so that it turned into a long piece of up to two million base pairs.
The chip works by placing the DNA in the centre of a glass plate, after which a flow of fluid stretches out the DNA.
This has been done successfully before, but not to the same degree.
“Previous techniques have only managed to stretch the DNA around 40 to 50 percent. Here, the DNA can still vibrate, which makes it difficult to take sharp photographs of it. With our nanofluidic chip, the DNA is stretched out completely, and that enables us to take significantly better photos of the DNA and see which areas light up,” says Pedersen.
DNA photo goes through database
The photo of the fluorescent DNA is then fed to a computer which compares it with an estimated light intensity profile of a reference DNA to see what it ought to look like.
This enables the researchers to determine whether there are greater genetic differences between the DNA they are studying and the reference DNA.
They can for instance see whether some pieces of the DNA have changed direction, as is frequently observed in cancer cells, or whether pieces of the DNA have been transferred from other chromosomes.
“The new technique also gives us a clearer idea of consecutive repetitions in the DNA’s base sequence. These repetitions occur frequently in DNA, and they can be difficult to detect when you only look at DNA pieces with a length of 300 base pairs,” says Pedersen.
“Here, you can often end up with many pieces of identical DNA, so you cannot say whether the DNA contains five, six or more of the repeated sequences. With the new technique, we can do that, since we study much longer pieces of DNA.”
New technique, new possibilities
Now that the researchers can study DNA molecules individually, they can study genetic differences between individual cells, e.g. cancer cells.
But the technique also makes it possible to study hitherto unknown pieces of DNA, where a high degree of repeated sequences of base pairs makes it impossible to examine them with conventional sequencing techniques.
One of these is the centromere, which is an area with a very high concentration of DNA at the centre of the chromosome.
”We can now investigate the general appearance of the DNA in the centromere,” he says. “The new technique does not, however, enable us to look at the sequence of individual base pairs as normal sequencing techniques can. But the new technique presents us with yet another tool to help us better understand how DNA is constructed.”
The scientific article also demonstrates that the DNA is not damaged by being dyed, stretched and photographed, and that it can subsequently be used for further analysis, e.g. normal DNA sequencing.
“It’s unique to be able to combine two analysis methods on a single copy of the genome. This means that our optical mapping can be developed into a method for preparing DNA samples for sequencing,” says Rodolphe Marie.
------------------------------
Read the Danish version of this article at videnskab.dk
Translated by: Dann Vinther