Researchers' Zone:
The brain acts like boiled spaghetti - a trait that can help us better understand brain diseases
New images of the brain's communication cables is a step on the way to earlier diagnosis of brain diseases and tailor made, patient-specific treatment
The brain controls everything we do, from fundamental processes like breathing and blinking, to more advanced actions like dancing or solving crosswords.
It consists of an extremely complex network of cells and communication cables called axons that send information between different parts of the brain and body.
When brain diseases such as Multiple Sclerosis and Alzheimer’s attack this network, they weaken and break down important brain functions like the ability to speak, move and remember.
To be able to develop cures for such diseases, we first need to understand exactly how they attack the brain tissue. This includes studying how the axons (brain connections) – which have for over a century been described as well-aligned, straight cylinders– are affected.
In our study, we found that the description of axons as cylinders is wrong, and this drastically changes the way we approach monitoring – and ultimately developing treatments for – disease.
The brain and its communication cables are super flexible
Just like with our muscles, stimulating the brain can improve how it works; the brain is able to reorganise its structure, function and connections.
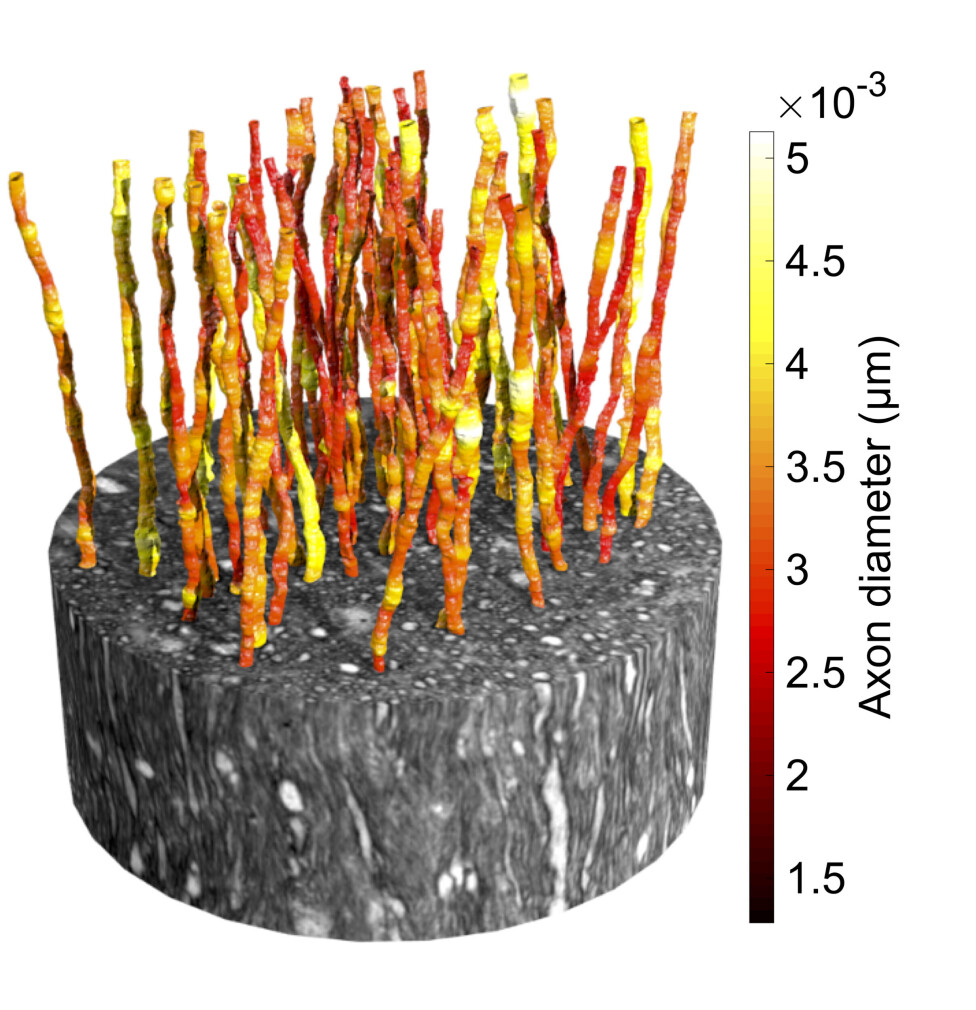
This flexibility of the brain also makes it robust to some diseases and injuries, as it is able to minimise their damage. By capturing ultra-high resolution 3D X-ray images of healthy brain tissue, we showed that the structure of the axons is different to what was previously believed and that the flexibility of the brain extends to the axons themselves.
In fact, one can think of axons as the soft, deformable spaghetti in a spaghetti-and-meatballs dish. They stick together in bundles, but are pushed and deformed around other structures in the brain tissue such as cell bodies and blood vessels.
Moreover, contrary to what was known before, the axons have varying diameters instead of a single, fixed size, as is also found in other recent studies using electron microscopy – another imaging technique.
We know that the speed with which signals are sent along brain connections depends on the diameters of the axons; the observed flexibility in axonal shape and diameter may thus have some impact on signal speed, but we do not yet know how much.
When diseases attack the brain network
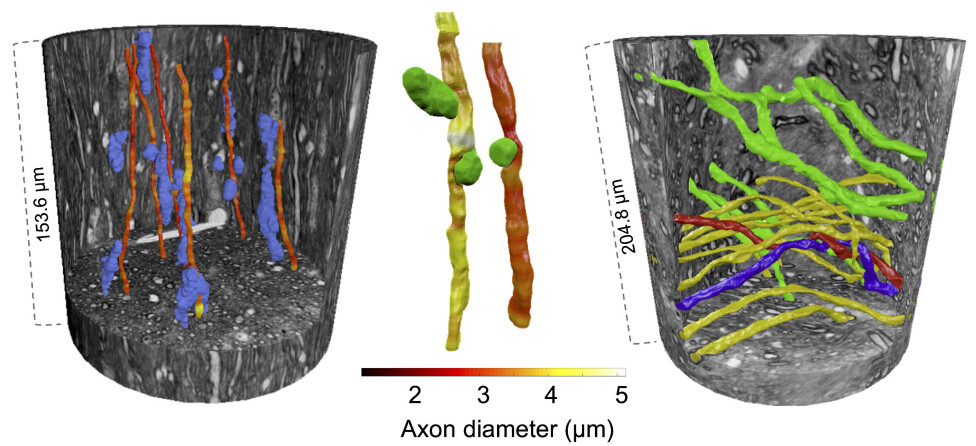
When different diseases attack, the brain employs its own healthcare system. This may involve, for example, an influx of helper cells that commute to the damaged region from different parts of the brain – just like ambulances responding to a car crash.
To reach the damaged site, these cells need to move along and between the tightly packed axons. In doing so, they end up pushing and deforming the axons when they pass by. It is possible that the flexibility in axon shape that we observe makes it possible for the helper cells to more efficiently reach and work on the damaged tissue.
In other 3D X-ray images that we are working with now, we can actually see how the helper cells accumulate around damaged brain connections.
Imaging the brain network
Images were captured at the European Synchrotron in Grenoble, the world’s leading source of X-rays. The synchrotron produces high energy X-rays that are far more intense than in regular medical X-ray scanners used to image e.g. broken legs.
Each 3D X-ray image contained an enormous amount of data – similar to the data required to stream a 16 hour movie. After analysing these images, the axons could be reconstructed and shown in 3D.
The 3D reconstructions reveal how axons change shape and bend around cells, blood vessels and even each other.
But there is a drawback: The 3D X-ray synchrotron imaging cannot be used on living people, and is instead limited to small tissue samples of less than a millimetre in size.
Therefore, it cannot be used to diagnose disorders in patients. Luckily, magnetic resonance imaging (MRI) can be used to study the living brain. More specifically, diffusion MRI can be used to study the anatomy at the microscopic scale – just like the 3D X-ray imaging.
There are significant differences between the techniques, however.
Firstly, the 3D X-ray images are obtained with light at such high energies that it is dangerous, whereas the MRI images are obtained with magnetic fields that have no known health effects.
Secondly, the 3D X-ray images are captured in very high nano-scale resolution and show single axons and cells in detail, but the resolution of MRI is in the millimetre range and cannot ‘see’ these structures.
With a special type of MRI, named diffusion MRI, we can detect the same details in the MRI scans as those we can directly see in the 3D X-ray images despite the poor image resolution.
Diffusion MRI uses magnetic fields to track the movement of water molecules within axons and cells. This movement of the water molecules can tell us about the size and organisation of axons in different parts of the brain.
So how can we detect these details in the MRI images without being able to actually see them?
To do this, we use a set of mathematical equations that describe what we expect the brain tissue environment to look like to build a virtual microscopic image of the tissue. However, this virtual image does not always match what we can see under the microscope and cannot be used as a reliable diagnostic tool.
Towards accurate and early diagnosis of brain diseases
One of today’s big imaging challenges is to be able to see how different diseases attack the brain tissue of patients, right from the very early stages of disease.
The brain’s incredible flexibility and ability to repair itself means that some diseases can ‘hide out’ in the brain, with long periods of time passing before the patient experiences symptoms.
MRI has the potential to catch early signs of the disease, but first the virtual image it produces needs to match what the anatomy truly looks like.
If we now instead use the new knowledge from the 3D X-ray images about the spaghetti-like shapes of the axons to tune the set of mathematical equations that describe the brain tissue environment in diffusion MRI, we get a more accurate virtual microscopic image and can better see what is truly going on in the brain.
This will mean that doctors can make more accurate – and hopefully earlier – diagnoses based on the MRI scans.
Furthermore, this new method would provide new knowledge on how diseases actually attack the brain tissue with time, potentially contributing to the development of new treatments as we can better follow how effective different treatments are.
Since the virtual microscopic image that is produced is unique to each patient, it is also possible that patient-specific treatment plans could be developed.
The research was performed as part of the RegionH-funded MAX4Imagers initiative that brings together researchers from the Technical University of Denmark, Copenhagen University, MAXIV synchrotron facility, Rigshospitalet and Hvidovre Hospital.
References:
- Mariam Andersson's profile (Region H)
- Tim Dyrby's profile (DTU)
- 'Axon morphology is modulated by the local environment and impacts the noninvasive investigation of its structure–function relationship', Proceedings of the National Academy of Sciences of the United States of America (2020), DOI: 10.1073/pnas.2012533117
- 'Does powder averaging remove dispersion bias in diffusion MRI diameter estimates within real 3D axonal architectures?', NeuroImage (2022), DOI: 10.1016/j.neuroimage.2021.118718
- 'Along-axon diameter variation and axonal orientation dispersion revealed with 3D electron microscopy: implications for quantifying brain white matter microstructure with histology and diffusion MRI', Brain Struct Funct (2019), DOI: 10.1007/s00429-019-018
- 'Automated 3D Axonal Morphometry of White Matter', Scientific Report (2019), DOI: 10.1038/s41598-019-42648-2
- 'Along-axon diameter variation and axonal orientation dispersion revealed with 3D electron microscopy: implications for quantifying brain white matter microstructure with histology and diffusion MRI', Brain Structure and Function (2019), DOI: 10.1007/s004
This article was originally published on our Danish sister site Forskerzonen.



